· Zhou, Tongqing et al. “Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.” Science (New York, N.Y.) vol. 376,6591 (2022): eabn8897. doi:10.1126/science.abn8897
· Zhao, Xiang et al. “Tuning T cell receptor sensitivity through catch bond engineering.” Science (New York, N.Y.) vol. 376,6589 (2022): eabl5282. doi:10.1126/science.abl5282
· Leggat, David J et al. “Vaccination induces HIV broadly neutralizing antibody precursors in humans.” Science (New York, N.Y.) vol. 378,6623 (2022): eadd6502. doi:10.1126/science.add6502
· Buchanan, Charles J et al. “Pathogen-sugar interactions revealed by universal saturation transfer analysis.” Science (New York, N.Y.) vol. 377,6604 (2022): eabm3125. doi:10.1126/science.abm3125
· Nabel, Katherine G et al. “Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.” Science (New York, N.Y.) vol. 375,6578 (2022): eabl6251. doi:10.1126/science.abl6251
· McCallum, Matthew et al. “Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement.” Science (New York, N.Y.) vol. 375,6583 (2022): 864-868. doi:10.1126/science.abn8652
· Liu, Kun-Hsiang et al. “NIN-like protein 7 transcription factor is a plant nitrate sensor.” Science (New York, N.Y.) vol. 377,6613 (2022): 1419-1425. doi:10.1126/science.add1104
· Kato, H et al. “Recognition of pathogen-derived sphingolipids in Arabidopsis.” Science (New York, N.Y.) vol. 376,6595 (2022): 857-860. doi:10.1126/science.abn0650
· Wang, Jue et al. “Scaffolding protein functional sites using deep learning.” Science (New York, N.Y.) vol. 377,6604 (2022): 387-394. doi:10.1126/science.abn2100
· Mannar, Dhiraj et al. “SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.” Science (New York, N.Y.) vol. 375,6582 (2022): 760-764. doi:10.1126/science.abn7760
· Bowen, John E et al. “Omicron spike function and neutralizing activity elicited by a comprehensive panel of vaccines.” Science (New York, N.Y.) vol. 377,6608 (2022): 890-894. doi:10.1126/science.abq0203
· Marklund, Emil et al. “Sequence specificity in DNA binding is mainly governed by association.” Science (New York, N.Y.) vol. 375,6579 (2022): 442-445. doi:10.1126/science.abg7427
· Reincke, S Momsen et al. “SARS-CoV-2 Beta variant infection elicits potent lineage-specific and cross-reactive antibodies.” Science (New York, N.Y.) vol. 375,6582 (2022): 782-787. doi:10.1126/science.abm5835
· Park, Young-Jun et al. “Imprinted antibody responses against SARS-CoV-2 Omicron sublineages.” Science (New York, N.Y.) vol. 378,6620 (2022): 619-627. doi:10.1126/science.adc9127
· Park, Young-Jun et al. “Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry.” Science (New York, N.Y.) vol. 375,6579 (2022): 449-454. doi:10.1126/science.abm8143
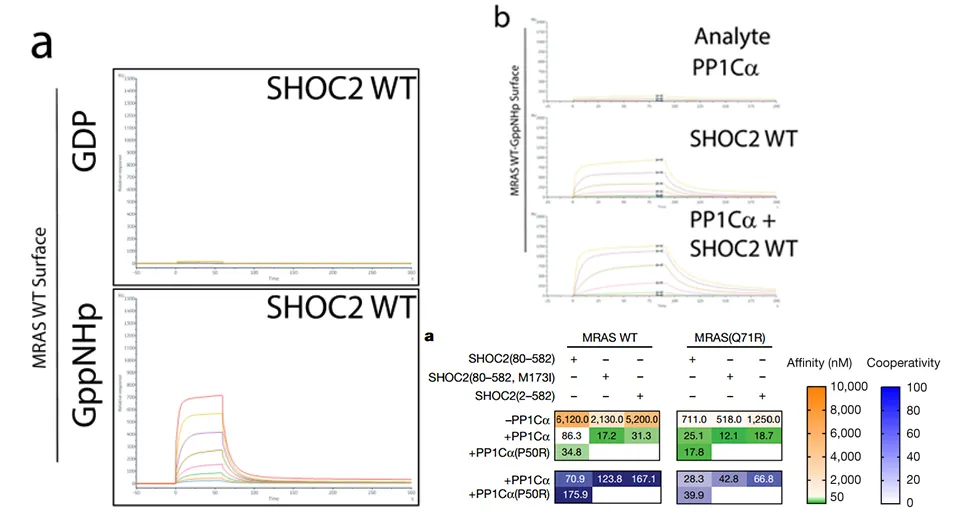
· Hauseman, Zachary J et al. “Structure of the MRAS-SHOC2-PP1C phosphatase complex.” Nature vol. 609,7926 (2022): 416-423. doi:10.1038/s41586-022-05086-1
· Wilson, Steven C et al. “Organizing structural principles of the IL-17 ligand-receptor axis.” Nature vol. 609,7927 (2022): 622-629. doi:10.1038/s41586-022-05116-y
· Lee, Jeong Hyun et al. “Long-primed germinal centres with enduring affinity maturation and clonal migration.” Nature vol. 609,7929 (2022): 998-1004. doi:10.1038/s41586-022-05216-9
· Marei, Hadir et al. “Antibody targeting of E3 ubiquitin ligases for receptor degradation.” Nature vol. 610,7930 (2022): 182-189. doi:10.1038/s41586-022-05235-6
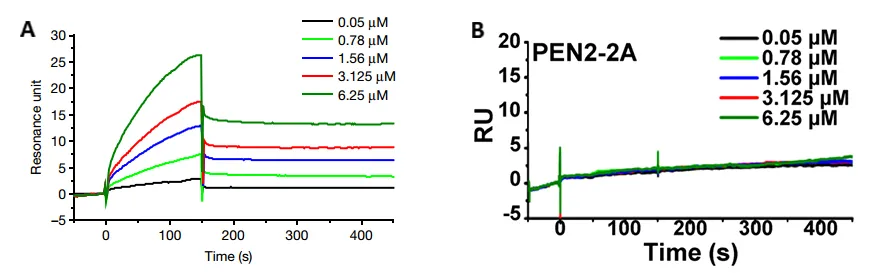
· Ma, Teng et al. “Low-dose metformin targets the lysosomal AMPK pathway through PEN2.” Nature vol. 603,7899 (2022): 159-165. doi:10.1038/s41586-022-04431-8
· Cao, Yunlong et al. “BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.” Nature vol. 608,7923 (2022): 593-602. doi:10.1038/s41586-022-04980-y
· Yang, Zhisen et al. “Structural insights into auxin recognition and efflux by Arabidopsis PIN1.” Nature vol. 609,7927 (2022): 611-615. doi:10.1038/s41586-022-05143-9
· Su, Nannan et al. “Structures and mechanisms of the Arabidopsis auxin transporter PIN3.” Nature vol. 609,7927 (2022): 616-621. doi:10.1038/s41586-022-05142-w
· Eichel, Kelsie et al. “Endocytosis in the axon initial segment maintains neuronal polarity.” Nature vol. 609,7925 (2022): 128-135. doi:10.1038/s41586-022-05074-5
· Wang, Qian et al. “Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5.” Nature vol. 608,7923 (2022): 603-608. doi:10.1038/s41586-022-05053-w
· Hochheiser, Inga V et al. “Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3.” Nature vol. 604,7904 (2022): 184-189. doi:10.1038/s41586-022-04467-w
· Shilts, Jarrod et al. “A physical wiring diagram for the human immune system.” Nature vol. 608,7922 (2022): 397-404. doi:10.1038/s41586-022-05028-x
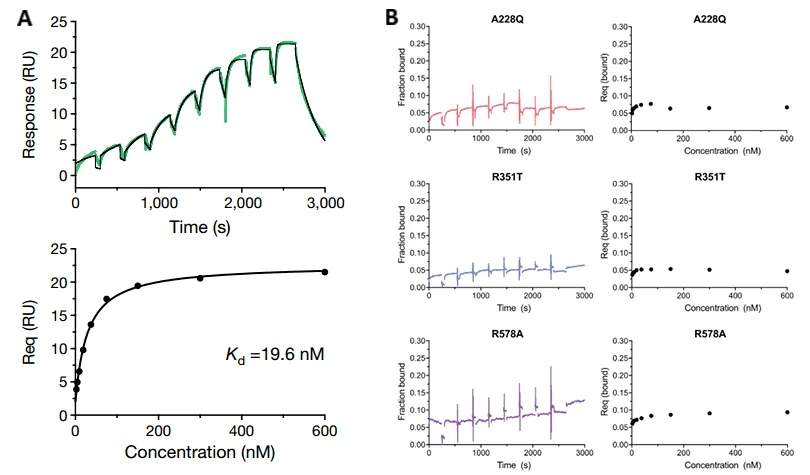
· Liau, Nicholas P D et al. “Structural basis for SHOC2 modulation of RAS signalling.” Nature vol. 609,7926 (2022): 400-407. doi:10.1038/s41586-022-04838-3
· Liu, Zunyong et al. “Phytocytokine signalling reopens stomata in plant immunity and water loss.” Nature vol. 605,7909 (2022): 332-339. doi:10.1038/s41586-022-04684-3
· Kim, Wooseob et al. “Germinal centre-driven maturation of B cell response to mRNA vaccination.” Nature vol. 604,7904 (2022): 141-145. doi:10.1038/s41586-022-04527-1
· Hu, Chun et al. “Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.” Nature vol. 602,7897 (2022): 518-522. doi:10.1038/s41586-021-04393-3
· Paik, Donggi et al. “Human gut bacteria produce ΤΗ17-modulating bile acid metabolites.” Nature vol. 603,7903 (2022): 907-912. doi:10.1038/s41586-022-04480-z
· Kwon, Diana. “Light-based sensors set to revolutionize on-site testing.” Nature vol. 607,7920 (2022): 834-836. doi:10.1038/d41586-022-02043-w
· Seung, Edward et al. “A trispecific antibody targeting HER2 and T cells inhibits breast cancer growth via CD4 cells.” Nature vol. 603,7900 (2022): 328-334. doi:10.1038/s41586-022-04439-0
· Yang, X., Garner, L.I., Zvyagin, I.V. et al. Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides. Nature 612, 771–777 (2022). https://doi.org/10.1038/s41586-022-05501-7
· Xu, Kun et al. “Protective prototype-Beta and Delta-Omicron chimeric RBD-dimer vaccines against SARS-CoV-2.” Cell vol. 185,13 (2022): 2265-2278.e14. doi:10.1016/j.cell.2022.04.029
· Li, Linjie et al. “Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.” Cell vol. 185,16 (2022): 2952-2960.e10. doi:10.1016/j.cell.2022.06.023
· Tuekprakhon, Aekkachai et al. “Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum.” Cell vol. 185,14 (2022): 2422-2433.e13. doi:10.1016/j.cell.2022.06.005
· Han, Pengcheng et al. “Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2.” Cell vol. 185,4 (2022): 630-640.e10. doi:10.1016/j.cell.2022.01.001
· Nutalai, Rungtiwa et al. “Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.” Cell vol. 185,12 (2022): 2116-2131.e18. doi:10.1016/j.cell.2022.05.014
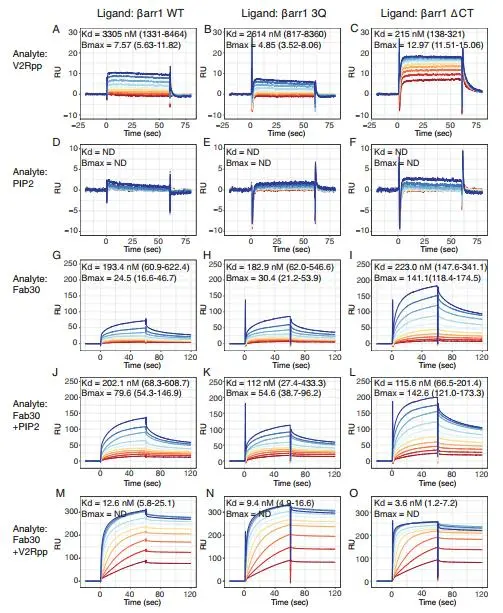
· Janetzko, John et al. “Membrane phosphoinositides regulate GPCR-β-arrestin complex assembly and dynamics.” Cell vol. 185,24 (2022): 4560-4573.e19. doi:10.1016/j.cell.2022.10.018
· Gagne, Matthew et al. “mRNA-1273 or mRNA-Omicron boost in vaccinated macaques elicits similar B cell expansion, neutralizing responses, and protection from Omicron.” Cell vol. 185,9 (2022): 1556-1571.e18. doi:10.1016/j.cell.2022.03.038
· Gagne, Matthew et al. “Protection from SARS-CoV-2 Delta one year after mRNA-1273 vaccination in rhesus macaques coincides with anamnestic antibody response in the lung.” Cell vol. 185,1 (2022): 113-130.e15. doi:10.1016/j.cell.2021.12.002
· Cui, Zhen et al. “Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.” Cell vol. 185,5 (2022): 860-871.e13. doi:10.1016/j.cell.2022.01.019
· Dejnirattisai, Wanwisa et al. “SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.” Cell vol. 185,3 (2022): 467-484.e15. doi:10.1016/j.cell.2021.12.046
· Akkermans, Onno et al. “GPC3-Unc5 receptor complex structure and role in cell migration.” Cell vol. 185,21 (2022): 3931-3949.e26. doi:10.1016/j.cell.2022.09.025
· Griffin, Matthew E, and Linda C Hsieh-Wilson. “Tools for mammalian glycoscience research.” Cell vol. 185,15 (2022): 2657-2677. doi:10.1016/j.cell.2022.06.016
· Wang, Qian et al. “Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants.” Cell, S0092-8674(22)01531-8. 14 Dec. 2022.

 收藏
收藏 询价
询价